Introduction
The interest in genetic modification of domestic animals is largely due to the possibility of using animals as bioreactors to produce pharmaceutical proteins, such as coagulation factors (Su et al., 2015), albumin (Sheng et al,. 2016), fibroblast growth factor 2 (FGF2) (Jeon et al., 2016), and others. Transgenic bovine production could benefit from somatic cell nuclear transfer (SCNT); however, such technology still presents low efficiencies of about 1-3% (Kues and Niemann, 2011). Cell type (Cho et al., 2004) and transgene introduction method (Cao et al., 2010) are important aspects that influence the efficiency of transgenic animal production by SCNT.
Fetal and adult fibroblasts are the cell types most commonly used in SCNT probably because these cells are easy to obtain, cultivate, and may undergo many cell divisions before reaching senescence (Bressan et al., 2008). Arat et al. (2001) showed for the first time the possibility of using granulosa cells in transgenic bovine production. Since then, different cell types, both fetal and adult, have been used as donor cells with variable efficiency (Gong et al., 2004; Feng et al., 2015). Hence, it is possible that more open chromatin configurations, such as those in fetal cells, are beneficial to transgenic production.
Gene transfer methods can be classified into transient or permanent or stable. Retroviral transduction is an example of permanent gene transfer because retroviruses use their infection machinery to integrate the transgene in a stable manner into the genome of the host cell. Using lipofection, a transient method, once inside the cell the transgene becomes dependent on cellular mechanisms to penetrate the nucleus and eventually integrate into chromosomal DNA. This process occurs spontaneously but rarely, and the integration occurs randomly, which makes the expression, when present, unpredictable, possibly causing low-efficiency transgenesis (Keravala and Calos, 2008).
There have been recent advances in the field of transgenesis, specifically endonucleases, mainly clustered regulatory interspaced short palindromic repeats (CRISPR-Cas9), which cleaves the DNA in a site-specific manner enabling to integrate a desired transgene with relatively high transgenesis efficiency (Hsu et al., 2014). Nevertheless, new technologies are still dependent on essential cellular and molecular mechanisms such as choice of cell type and the most appropriate gene transfer method. Managing these check points should improve transgenic results.
Thus, this study aimed to investigate the effect of cell type (fetal or adult fibroblasts, and cumulus cells) and gene transfer method (lipofection or lentiviral transduction) on gene incorporation, expression, and fluorescence intensity of transgene on bovine cells analyzed by flow cytometry.
Materials and methods
Ethical considerations
All cell types used in this work were obtained from bovine tissues collected from a slaughterhouse in Campos dos Goytacazes, Brazil.
Cell culture establishment
Fetal fibroblasts were obtained from a tissue fragment culture from the dorsal skin of 50-day fetuses, and adult fibroblasts from a subcutaneous tissue fragment culture of adult cattle ear. Collected tissues were cut into small pieces and cultured in TCM without HEPES with 10% fetal bovine serum, and 1% (v/v) penicillin/streptomycin (10 000 U/ml penicillin G, 10 000 mg/ml streptomycin) culture medium at 38.5°C; these were placed in 35-mm tissue culture plates and kept in a humidified atmosphere of 5% CO2 in air until 80% confluence, approximately, was reached.
A cumulus cell culture was established by aspirating ovarian antral follicles from slaughterhouse ovaries. The follicles were transferred into 35-mm tissue culture plates containing culture medium and then cultured for at least 24 hours. The oocytes were then removed, and the cumulus cells were cultured until they reached 80% confluence, approximately. The cultures from different cell types were then tripsinized with 0.25% trypsin. After two passages, the cells were frozen in TCM without HEPES, supplemented with 40% FBS and 10% dimethyl sulfoxide. Experiments were conducted with cell lines at the same or very similar (<5) passages.
Transgenic cell establishment
The cells were submitted to two different gene transfer methods: lentiviral transduction (Group 1, G1), and lipofection (Group 2, G2). Lentiviral transduction (G1) was performed with lentiviral particles following the ViraPower lentiviral expression system (Life Technologies, Carlsbad, California, USA) protocol, and the FUGW (flap- Ub promoter-GFP-WRE) plasmid containing the reporter enhanced Green Fluorescent Protein (eGFP) gene under the control of Ubiquitin C promoter (constitutive expression) (Lois et al., 2002). For this, 293FT cells (Invitrogen, Carlsbad, California, USA) were transfected using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, Massachusetts, USA) with 1,2 µg pLP1, and pLP2; 2,4µg pLP/ VSVG plasmids, and 1 µg FUGW for each well of a six-well plate, in accordance to the manufacturer’s instructions. Twenty-four and 48 hours after 293FT cell transfection, supernatant culture medium containing lentiviral particles was collected, filtered, and deposited on the wells containing the different cell types (1mL/well).
Lipofection (G2) was carried out using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to the manufacturer’s instructions such that each well contained 1 µg of FUGW plasmid. The control group was not subjected to any method of transfection. Transduction and lipofectation were performed in 6-well plates.
Fluorescence evaluation of the post-transfected cells by flow cytometry
Flow cytometry analysis was carried out 48 hours post-transfection using BD FACSAria flow cytometer and FACSDiva software (BD Biosciences, San Jose, CA, USA).
Fluorescence was excited with a 488-nm laser, and read using a 530/30-nm filter. The different cell types were identified and selected from debris by size (forward-scatter light (FSC-H)) and complexity (side- scattered light (SSC-H)) analyses. Two variables were measured by cytometric analysis: (i) the transfection efficiency (Green Fluorescent Protein positive cells - GFP+ cells, read by fluorescein isothiocyanate -FITC- channel), and (ii) the intensity of fluorescence emitted by GFP+ cells (means of arbitrary units, FACSDiva software, BD Biosciences, San Jose, CA, USA).
Morphological evaluation of the post-transfection cells observed under light microscopy
The cells submitted to transduction or lipofection were analyzed through optical microscopy (200X) (TE300, Nikon, Osaka, Japan) 24 hours post- transfection for overall viability analysis. The presence of cellular debris, detached cells, and cytoplasmic vacuoles -considered as cell death indicators- was visually assessed.
Statistical analysis
After data consistency and descriptive statistics analysis (mean, standard deviation, and coefficient of variation), analysis of variance was used to verify differences between the cell types that underwent different treatments, and to determine if there was any interaction between the two variables. The interaction was considered significant at p<0.05, and analyses were done for each cell type subjected to each treatment. The means were compared by the Student- Newman-Keuls (SNK) test of SAS® software, version 6.03, 1988 (SAS Institute Inc., Cary, NC, USA).
Results
Cell type influences the transfection method efficiency
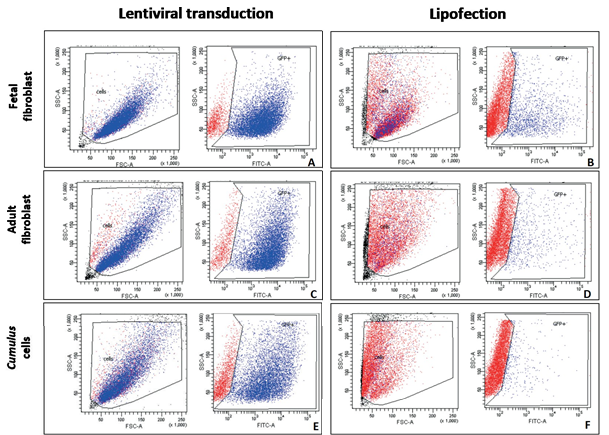
The lentiviral system was the most efficient gene transfer method (based on the percentage of cells positive for green fluorescence, GFP+) regardless of the cell type used, with a higher number of positive cells per sample (FF 88.8% ± 0.98; AF 91.6% ± 2.96; and CC 60.7% ± 14.7, p<0.05). Among the cell types examined, CC showed the lowest (p<0.05) transgene expression levels, and differed from the others. The use of lipofection resulted in a significantly lower (p<0.05) transfection percentage than when the lentiviral system was applied. Fetal fibroblasts presented significantly higher (p<0.05) transfection levels (17.8% ± 2.82), followed by AF (10.66% ± 0.65) and, finally, by CC (3.9% ± 1.97) (Table 1 and Figure 1).
Table 1
Percentage of fetal fibroblasts (FF), adult fibroblasts (AF), and cumulus cells (CC) emitting green fluorescence (GFP+) after lentiviral transduction or lipofection to introduce the FUGW plasmid, as analyzed by flow cytometry.
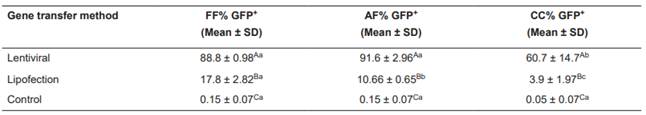
Figure 1
Gene transfer efficiency measured by the number of GFP+ cells (positive cells on FITC channel in FACSAria cytometer) analyzed by flow cytometry. Blue dots refer to cells positive for GFP expression and red dots refer to negative cells. The effects of lentiviral transduction and lipofection, and cell type (rows) on transgene expression in bovine cells were compared. Fluorescence was excited with a 488-nm laser, and read using a 530/30-nm filter. A and B: dotplots for fetal fibroblasts identification (selection FSC-A (size) x SSC-A (complexity)), and fluorescence (SSC-A x FITC-A) after lentiviral transduction and lipofection, respectively. C and D: dotplots for cell identification and fluorescence for lentiviral transductions and lipofection for adult fibroblasts, and E and F: dotplots for cell identification and fluorescence for lentiviral transductions and lipofection for cumulus cells.
Fluorescence intensity, which is related to the expression level of the GFP protein, did not differ significantly (p<0.05) between lentiviral transduction and lipofection, regardless of the cell type analyzed.
In both treatments, adult fibroblasts showed higher transgene expression levels, which did not differ from those of cumulus cells, but did differ from the fetal fibroblasts (p<0.05) (Table 2).
Morphologic evaluation of the post-transfection cells
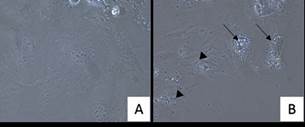
At visual assessment, cells submitted to lipofection presented high number of dead cells (detaching debris) per field and presence of cytoplasmic vacuoles. Cumulus cell cultures submitted to Lipofectamine showed the greatest signs of cellular death, with high presence of cytoplasmic vacuoles, cellular debris, and detached cells (Figure 2).
Discussion
The low efficiency of transgenic animal production by SCNT can be attributed to many factors, including the transfection method and the differentiation stage of the donor cell. This study compared the effects of two gene transfer methods, lentiviral transduction and lipofection, and of cell type (AF, FF, and CC) on transgene expression efficiency.
Lentiviral transduction was the most efficient method of gene transfer, differing significantly from lipofection regardless of cell type used and resulting in a high number of GFP+ cells per sample. Retroviral vectors are a powerful genetic tool for generation of cells for transgenic animal production, mainly due to the fact that retroviruses use their own biological infection mechanisms to achieve cell transduction, and DNA integration in a stable and lasting method.
The present study used lentiviral vectors, which are able to infect cells which are or are not in division and have the ability to carry out their pre-integration complex in an active manner into the host cell, ensuring DNA integration (Denning et al., 2013). Probably, these factors resulted in the best transgene expression rates when the lentiviral system was used.
On the other hand, when an exogenous DNAsequence is inserted into a mammalian cell through lipofection, the DNA is released into the cell and becomes dependent on the cellular machinery to penetrate the nucleus. The integration into chromosomal DNA is a spontaneous and rare event, which makes the expression, when present, unpredictable (Keravala and Calos, 2008). This fact can explain the lower transfection rates obtained when lipofection was used. Moreover, the use of lipofection has been generally associated with transient transfection (Ooi et al., 2016; Fuge et al., 2017), a fact that limits its application in the production of transgenic animals (Bressan et al., 2008).
Good transfection results have been obtained using lipofection in cellular lineages, such as 293 and HELA cells (Thomas and Smart, 2005; Dalton and Barton, 2014; Vink et al., 2014); however, gene transfer into primary culture cells is still limited. Lipofection is a simple and nonviral methodology (Wang et al., 2015), nevertheless, improvement of protocols is needed for individual laboratory conditions, gene construction and type of cell to increase transfection rates and to reduce cell death post-transfection, which was expressive in this study when this transfection method was used.
Cao et al. (2010) compared the gene transfer efficiency (pUb-eGFP-Fluc) of different gene transfer methods (electroporation, lipofection, nucleofection, and lentiviral transduction) in human embryonic stem cells. They reported that the gene transfer efficiencies of lentiviral transduction and nucleofection were about 25% higher than those of lipofection and electroporation.
A study using human periodontal ligament stem cells (hPDLSCs) compared transfection efficiency of five nonviral-gene-transfer methods [lipofection using Lipofectamine 2000, polyethylenimine, GBfectene- Elite transfection reagent, X-tremeGENE HP DNA Transfection Reagent, and Magnet-Assisted Transfection (MATra)] and lentiviral vectors using fluorescence microscopy and flow cytometry. According to the authors, MATra was the most effective nonviral method reaching around 11% of transfection, while, the others four nonviral methods, including lipofection, resulted in less than 6% efficiency. When lentiviral vectors were used, transduction reached about 95% success. Furthermore, when lipofection reagents were used, the authors also reported cytotoxicity, irregular cell morphology and cells mortality, similar to those observed in the present study (Wang et al., 2015).
In the present study, using fetal fibroblasts, lipofection resulted in about 20% gene transfer, which was significantly higher than in adult cells subjected to the same treatment. These results may be attributed to cell differentiation status, which is related to the chromatin epigenetic configuration (Ng and Gurdon, 2008; Chen and Dent 2014). A more open chromatin configuration, such as that probably found in fetal cells, may be beneficial to the introduction of genes by lipofection, in which the integration to chromosomal DNA occurs spontaneously (Song et al., 2014).
Furthermore, transfection efficiency of lipofection is directly related to cell division rate (Gresch et al., 2004). In the present work, fetal fibroblasts reached confluence faster than adult cells (data not shown). Such factors can be attributed to the advantage of lipofection when fetal cells are used.
The lentiviral system is well known to result in random and, sometimes, multiple transgene integration events into the host cell DNA. Such fact can increase undesirable DNA integration into encoding sites (Zhang et al., 2012), a problematic issue because position effect variegation can profoundly affect transgene expression, leading to unpredictable transgene expression, including the disruption of endogenous genes, and phenotype (Beard et al., 2006; Rulicke and Hubscher, 2000; Soriano et al., 1987; Williams et al., 2008). Moreover, gene silencing by DNA methylation has been reported due to the presence of viral sequences (Hofmann, 2006). Therefore, previous characterization of cell lines prior to use as donor cells in SCNT is important to guarantee the welfare of transgenic animals (Bressan et al., 2011).
When retroviruses were used in our study, fluorescence intensity -which may be related to the copy number delivery into DNA cells (Soboleski et al., 2005)- did not significantly differ from that obtained with lipofection use; thus, no deleterious effect of retroviral transfection was observed.
To obtain the appropriate expression of a particular protein, the genetic code must be transcribed into the mRNA molecule. Toward this end, a transcription factor recognizes the gene promoter sequence and initiates the mRNA transcription molecule that is translated into the protein. The present work used the FUGW vector (Lois et al., 2002), which contains-besides the coding region for the reporter gene- the GPF; a regulatory posttranscriptional region of the woodchuck hepatitis virus (WRE), introduced to increase the protein transcription level. The human immunodeficiency virus-1 flap was inserted between LTRs to increase viral titers, which are part of the plasmid. Lois et al. (2002) used the human ubiquitin-C promoter in the vector construction. According to Lois et al. (2002), this promoter led to a better transgene expression on the cell types analyzed.
In the present study, it was expected that fetal fibroblasts would present better results since fetal cells are known to be more epigenetically plastic, due to its open chromatin status. However, interestingly, adult fibroblasts showed higher fluorescence intensity than fetal fibroblasts in both methods (Table 2). The selection of plasmid construction according to the type of cell seems to be essential for the production of transgenic cells, considering that some promoters are more efficiently expressed in certain tissues than in others (Zheng and Baum 2008), thus leading to different fluorescence levels in different cell types.
Despite the advances in this field, production of transgenic animals by SCNT is still considered a low-efficiency technique (Yang et al., 2008; Bertolini et al., 2016) mainly due to the high number of steps involved in the process from donor cell preparation to transgenic animal birth. The relationships between type and differentiation stage of cells, transfection method, and transgene expression level are not well defined. Further research is needed to understand these relations given that epigenetic factors may be involved and thus may influence the results. To the best of our knowledge, no previous reports or studies have evaluated the effects of transfection method and cell type on transfection rates and transgene expression in cattle. Manipulation of factors responsive for efficiency discrepancies could increase the efficiency of production of transgenic cattle by TN.
In conclusion, gene transfer efficiency differs between cell types depending on the transfection method used. Second, lentiviral transduction resulted in higher transfection rates regardless of cell type. Third, for laboratories without capacities for viral manipulation, lipofection may present an alternative for cell production that transiently expresses transgenes. Finally, although epigenetic profiles may vary between cell lines, and therefore it was expected that fetal lines would be more prone to present a better gene transfer rate, in the conditions of this study the best results were obtained using adult fibroblasts; this could represent an advantage for the production of transgenic animals by nuclear transfer, in which knowledge of animal genotype and phenotype is desired.