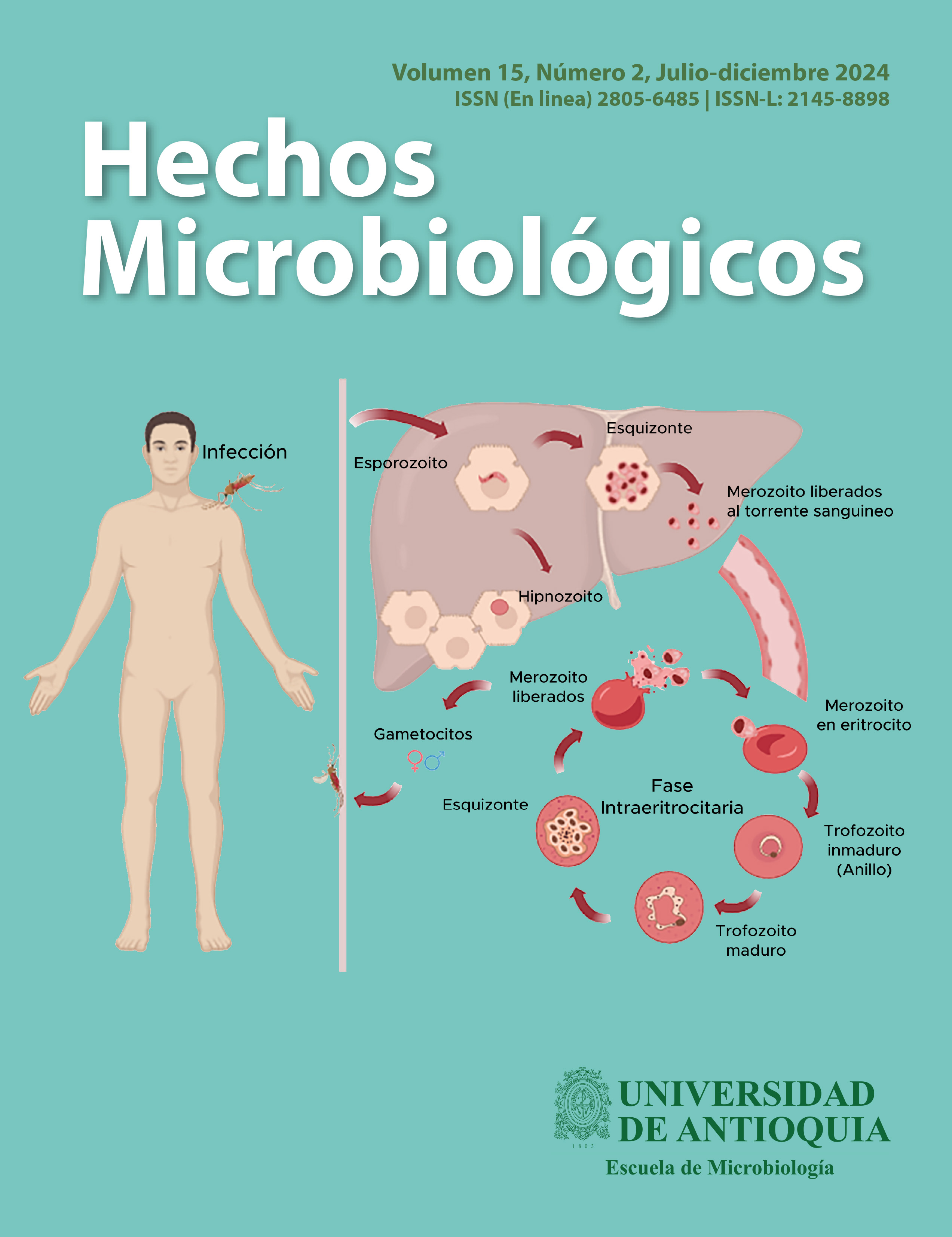
Explorando los mecanismos de invasión celular de Plasmodium Vivax a través del sistema de grupo sanguíneo Duffy
DOI:
https://doi.org/10.17533/udea.hm.v15n2a03Palabras clave:
DARC, Malaria, Plasmodium vivax, PvDBP, PvEBP, Sistema de grupo sanguíneo DuffyResumen
Introducción: La malaria por P. vivax representa un grave problema de salud pública a nivel mundial, especialmente en regiones tropicales. Se creía que la ausencia del antígeno Duffy protegía contra esta infección. Sin embargo, estudios han demostrado que P. vivax puede infectar a individuos Duffy negativos, sugiriendo mecanismos de invasión alternativos. Objetivo: Este artículo tiene como objetivo comprender los mecanismos de invasión que P. vivax utiliza para infectar eritrocitos en individuos Duffy positivos, así como explorar las posibles vías de infección en individuos Duffy negativos. Metodología: Búsqueda bibliográfica en las bases de datos PubMed, LILACS y Google Scholar, publicaciones entre los años 2009 al 2024, en idiomas español e inglés, empleando los siguientes términos: DARC, malaria, P. vivax, PvEBP, PvDBP y sistema de grupo sanguíneo Duffy. Resultados: La invasión por P. vivax se ha vinculado principalmente a la interacción entre la proteína PvDBP y DARC. Sin embargo, el creciente reporte de infecciones en individuos Duffy-negativos ha llevado al estudio de posibles rutas alternativas. Entre las hipótesis planteadas se incluyen la duplicación del gen PvDBP, la interacción entre TfR1 y PvRBP2b, y el PvEBP como ligando alternativo, debido a sus similitudes estructurales con PvDBP. Conclusión: La comprensión de los mecanismos de invasión de P. vivax en eritrocitos es importante para el desarrollo a futuro de terapias innovadoras y estrategias de control. La identificación de rutas alternativas, especialmente en individuos Duffy-negativos, destaca la necesidad de explorar nuevos blancos terapéuticos para interrumpir el ciclo sanguíneo del parásito y avanzar en su eliminación.
Descargas
Citas
Asua V, Tukwasibwe S, Conrad M, Walakira A, Nan-kabirwa JI, Mugenyi L, et al. Plasmodium Species Infecting Children Presenting with Malaria in Ugan-da. Am J Trop Med Hyg 2017;97:753–7. https://doi.org/10.4269/ajtmh.17-0345.
Barnwell JW, Nichols ME, Rubinstein P. In vitro evaluation of the role of the Duffy blood group in erythrocyte invasion by Plasmodium vivax. J Exp Med 1989;169:1795–802. https://doi.org/10.1084/jem.169.5.1795.
Bermúdez M, Moreno-Pérez DA, Arévalo-Pinzón G, Curtidor H, Patarroyo MA. Plasmodium vivax in vitro continuous culture: The spoke in the wheel. Malar J 2018;17. https://doi.org/10.1186/s12936-018-2456-5.
Bouyssou I, Martínez FJ, Campagne P, Ma L, Doder-er-Lang C, Chitnis CE, et al. Plasmodium vivax blood stage invasion pathways: Contribution of omics technologies in deciphering molecular and cellular mechanisms. C R Biol 2022;345:91–133. https://doi.org/10.5802/crbiol.95.
BRAY RS. The susceptibility of Liberians to the Mada-gascar strain of Plasmodium vivax. J Parasitol 1958;44. https://doi.org/10.2307/3274317.
Carvalho TA, Queiroz MG, Cardoso GL, Diniz IG, Sil-va AN, Pinto AY, et al. Plasmodium vivax infection in Anajás, State of Pará: no differential resistance profile among Duffy-negative and Duffy-positive individuals. Malar J 2012;11:430. https://doi.org/10.1186/1475-2875-11-430.
Chan LJ, Dietrich MH, Nguitragool W, Tham WH.Plasmodium vivax Reticulocyte Binding Proteins for invasion into reticulocytes. Cell Microbiol 2020;22. https://doi.org/10.1111/cmi.13110.
Culleton R, Carter R. African Plasmodium vivax: Distri-bution and origins. Int J Parasitol 2012;42. https://doi.org/10.1016/j.ijpara.2012.08.005.
Dayananda KK, Achur RN, Gowda DC. Epidemiol-ogy, drug resistance, and pathophysiology of Plasmo-dium vivax malaria. J Vector Borne Dis 2018;55. https://doi.org/10.4103/0972-9062.234620.
de Carvalho GB, de Carvalho GB. Duffy blood group system and the malaria adaptation process in humans. Rev Bras Hematol Hemoter 2011;33. https://doi.org/10.5581/1516-8484.20110016.
Dluzewski AR, Ling IT, Hopkins JM, Grainger M, Margos G, Mitchell GH, et al. Formation of the Food Vacuole in Plasmodium falciparum: A Potential Role for the 19 kDa Fragment of Merozoite Surface Pro-tein 1 (MSP119). PLoS One 2008;3:e3085. https://doi.org/10.1371/journal.pone.0003085.
Douglas NM, Anstey NM, Buffet PA, Poespoprodjo JR, Yeo TW, White NJ, et al. The anaemia of Plasmo-dium vivax malaria. Malar J 2012;11:135. https://doi.org/10.1186/1475-2875-11-135.
Ellis JM, Stubbs TH, Young MD. Some Characteristics of Foreign Vivax Malaria Induced in Neurosyphilitic Patients 1,2. Am J Trop Med Hyg 1947;s1-27:585–96. https://doi.org/10.4269/ajtmh.1947.s1-27.585.
Golassa L, Amenga-Etego L, Lo E, Amambua-Ngwa A. The biology of unconventional invasion of Duffy-negative reticulocytes by Plasmodium vivax and its im-plication in malaria epidemiology and public health. Malar J 2020;19. https://doi.org/10.1186/s12936-020-03372-9.
Gonzalez L, Vega J, Ramirez JL, Bedoya G, Car-mona-Fonseca J, Maestre A. Relationship between genotypes of the Duffy blood groups and malarial in-fection in different ethnic groups of Choco, Colombia. Colomb Med (Cali) 2012;43:189–95.
Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, et al. The global distribution of the Duffy blood group. Nat Commun 2011;2:266. https://doi.org/10.1038/ncomms1265.
INS-Boletin epidemiologico semanal. Semana epi-demiológica 52; 24 al 30 de diciembre de 2023. 2023.
Kadekoppala M, Holder AA. Merozoite surface pro-teins of the malaria parasite: The MSP1 complex and the MSP7 family. Int J Parasitol 2010;40:1155–61. https://doi.org/10.1016/j.ijpara.2010.04.008.
Kanjee U, Rangel GW, Clark MA, Duraisingh MT. Molecular and cellular interactions defining the tropism of Plasmodium vivax for reticulocytes. Curr Opin Microbiol 2018;46. https://doi.org/10.1016/j.mib.2018.10.002.
Kwiatkowski , Dominic P. How Malaria Has Affected the Human Genome and What Human Genetics Can Teach Us about Malaria . The American Journal of Hu-man Genetics n.d.;77.
Loy DE, Liu W, Li Y, Learn GH, Plenderleith LJ, Sun-dararaman SA, et al. Out of Africa: origins and evolu-tion of the human malaria parasites Plasmodium falci-parum and Plasmodium vivax. Int J Parasitol 2017;47. https://doi.org/10.1016/j.ijpara.2016.05.008.
Malleret B, Li A, Zhang R, Tan KSW, Suwanarusk R, Claser C, et al. Plasmodium vivax: restricted tropism and rapid remodeling of CD71-positive reticulocytes. Blood 2015;125:1314–24. https://doi.org/10.1182/blood-2014-08-596015.
Martínez Santander C ACCCSLCVHRAK. Mala-ria: Un problema de alta prevalencia en habitantes de regiones tropicales de Latinoamérica. Ateneo 2023;25(2):110–28.
Ménard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Mala-gasy people. Proc Natl Acad Sci U S A 2010;107:5967–71. https://doi.org/10.1073/pnas.0912496107.
Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de Sousa B, et al. Duffy Negative Antigen Is No Longer a Barrier to Plasmodium vivax – Molecular Evidences from the African West Coast (Angola and Equatorial Guinea). PLoS Negl Trop Dis 2011;5:e1192. https://doi.org/10.1371/journal.pntd.0001192.
Miller LH, Mason SJ, Clyde DF, McGinniss MH. The Resistance Factor to Plasmodium vivax in Blacks. The Duffy Blood Group Genotype, FyFy. N Engl J Med 1976;295.
Molina-Franky J, Reyes C, Picón Jaimes YA, Kalkum M, Patarroyo MA. The Black Box of Cellular and Mo-lecular Events of Plasmodium vivax Merozoite Invasion into Reticulocytes. Int J Mol Sci 2022;23. https://doi.org/10.3390/ijms232314528.
Molina-Franky J, Reyes C, Picón Jaimes YA, Kalkum M, Patarroyo MA. The Black Box of Cellular and Mo-lecular Events of Plasmodium vivax Merozoite Invasion into Reticulocytes. Int J Mol Sci 2022;23. https://doi.org/10.3390/ijms232314528.
Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, et al. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria para-site. Lancet Infect Dis 2009;9. https://doi.org/10.1016/S1473-3099(09)70177-X.
Niangaly A, Gunalan K, Ouattara A, Coulibaly D, Sá JM, Adams M, et al. Plasmodium vivax Infections over 3 Years in Duffy Blood Group Negative Malians in Bandiagara, Mali. American Journal of Tropical Medi-cine and Hygiene 2017;97. https://doi.org/10.4269/ajtmh.17-0254.
Organización Mundial de la Salud/ Organización Panamericana de la Salud. Malaria. Https://Www-WhoInt/Es/News-Room/Fact-Sheets/Detail/Malaria2023.
Pachebat JA, Kadekoppala M, Grainger M, Dlu-zewski AR, Gunaratne RS, Scott-Finnigan TJ, et al.Extensive proteolytic processing of the malaria para-site merozoite surface protein 7 during biosynthesis and parasite release from erythrocytes. Mol Biochem Parasitol 2007;151:59–69. https://doi.org/10.1016/j.molbiopara.2006.10.006.
Picón-Jaimes YA, Lozada-Martinez ID, Buelvas MCF, Sarmiento AFA, Baez GAS, Erazo DYN, et al. Evolution of Plasmodium vivax and resistance patterns for infection based on Duffy genotype and pheno-type. Infezioni in Medicina 2023;31:350–8. https://doi.org/10.53854/liim-3103-8.
Poirier P, Doderer-Lang C, Atchade PS, Lemoine J-P, de l’Isle M-LC, Abou-bacar A, et al. The hide and seek of Plasmodium vivax in West Africa: report from a large-scale study in Beninese asymptomatic subjects. Malar J 2016;15:570. https://doi.org/10.1186/s12936-016-1620-z.
Popovici J, Roesch C, Rougeron V. The enigmatic mechanisms by which Plasmodium vivax infects Duffy-negative individuals. PLoS Pathog 2020;16. https://doi.org/10.1371/journal.ppat.1008258.
Russo G, Faggioni G, Paganotti GM, Djeunang Dongho GB, Pomponi A, De Santis R, et al. Molecu-lar evidence of Plasmodium vivax infection in Duffy negative symptomatic individuals from Dschang, West Cameroon. Malar J 2017;16:74. https://doi.org/10.1186/s12936-017-1722-2.
Tyagi K, Hossain ME, Thakur V, Aggarwal P, Mal-hotra P, Mohmmed A, et al. Plasmodium vivax Tryptophan Rich Antigen PvTRAg36.6 Interacts with PvETRAMP and PvTRAg56.6 Interacts with PvMSP7 during Erythrocytic Stages of the Parasite. PLoS One 2016;11:e0151065. https://doi.org/10.1371/journal.pone.0151065.
Young MD, Eyles DE, Burgess RW, Jeffery GM. Ex-perimental Testing of the Immunity of Negroes to Plasmodium vivax. J Parasitol 1955;41:315. https://doi.org/10.2307/3274214.
Adams JH, Hudson DE, Torii M, Ward GE, Wellems TE, Aikawa M, et al. The duffy receptor family of plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell 1990;63:141–53. https://doi.org/10.1016/0092-8674(90)90295-P.
Asua V, Tukwasibwe S, Conrad M, Walakira A, Nankabirwa JI, Mugenyi L, et al. Plasmodium Species Infecting Children Presenting with Malaria in Uganda. Am J Trop Med Hyg 2017;97:753–7. https://doi.org/10.4269/ajtmh.17-0345.
Auburn S, Getachew S, Pearson RD, Amato R, Miotto O, Trimarsanto H, et al. Genomic Analysis of Plasmodium vivax in Southern Ethiopia Reveals Selective Pressures in Multiple Parasite Mechanisms. J Infect Dis 2019;220:1738–49. https://doi.org/10.1093/infdis/jiz016.
Barnwell JW, Nichols ME, Rubinstein P. In vitro evaluation of the role of the Duffy blood group in erythrocyte invasion by Plasmodium vivax. J Exp Med 1989;169:1795–802. https://doi.org/10.1084/jem.169.5.1795.
Bermúdez M, Moreno-Pérez DA, Arévalo-Pinzón G, Curtidor H, Patarroyo MA. Plasmodium vivax in vitro continuous culture: The spoke in the wheel. Malar J 2018;17. https://doi.org/10.1186/s12936-018-2456-5.
Bermúdez M, Moreno-Pérez DA, Arévalo-Pinzón G, Curtidor H, Patarroyo MA. Plasmodium vivax in vitro continuous culture: the spoke in the wheel. Malar J 2018;17:301. https://doi.org/10.1186/s12936-018-2456-5.
Bouyssou I, Martínez FJ, Campagne P, Ma L, Doderer-Lang C, Chitnis CE, et al. Plasmodium vivax blood stage invasion pathways: Contribution of omics technologies in deciphering molecular and cellular mechanisms. C R Biol 2022;345:91–133. https://doi.org/10.5802/crbiol.95.
BRAY RS. The susceptibility of Liberians to the Madagascar strain of Plasmodium vivax. J Parasitol 1958;44. https://doi.org/10.2307/3274317.
Carvalho TA, Queiroz MG, Cardoso GL, Diniz IG, Silva AN, Pinto AY, et al. Plasmodium vivax infection in Anajás, State of Pará: no differential resistance profile among Duffy-negative and Duffy-positive individuals. Malar J 2012;11:430. https://doi.org/10.1186/1475-2875-11-430.
Chan LJ, Dietrich MH, Nguitragool W, Tham WH. Plasmodium vivax Reticulocyte Binding Proteins for invasion into reticulocytes. Cell Microbiol 2020;22. https://doi.org/10.1111/cmi.13110.
Cheng Y, Lu F, Wang B, Li J, Han J-H, Ito D, et al. Plasmodium vivax GPI-anchored micronemal antigen (PvGAMA) binds human erythrocytes independent of Duffy antigen status. Sci Rep 2016;6:35581. https://doi.org/10.1038/srep35581.
Chitnis CE, Chaudhuri A, Horuk R, Pogo AO, Miller LH. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J Exp Med 1996;184:1531–6. https://doi.org/10.1084/jem.184.4.1531.
Chitnis CE, Miller LH. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med 1994;180:497–506. https://doi.org/10.1084/jem.180.2.497.
Choe H, Moore MJ, Owens CM, Wright PL, Vasilieva N, Li W, et al. Sulphated tyrosines mediate association of chemokines and Plasmodium vivax Duffy binding protein with the Duffy antigen/receptor for chemokines (DARC). Mol Microbiol 2005;55:1413–22. https://doi.org/10.1111/j.1365-2958.2004.04478.x.
Culleton R, Carter R. African Plasmodium vivax: Distribution and origins. Int J Parasitol 2012;42. https://doi.org/10.1016/j.ijpara.2012.08.005.
Dayananda KK, Achur RN, Gowda DC. Epidemiology, drug resistance, and pathophysiology of Plasmodium vivax malaria. J Vector Borne Dis 2018;55. https://doi.org/10.4103/0972-9062.234620.
de Carvalho GB, de Carvalho GB. Duffy blood group system and the malaria adaptation process in humans. Rev Bras Hematol Hemoter 2011;33. https://doi.org/10.5581/1516-8484.20110016.
Dluzewski AR, Ling IT, Hopkins JM, Grainger M, Margos G, Mitchell GH, et al. Formation of the Food Vacuole in Plasmodium falciparum: A Potential Role for the 19 kDa Fragment of Merozoite Surface Protein 1 (MSP119). PLoS One 2008;3:e3085. https://doi.org/10.1371/journal.pone.0003085.
Douglas NM, Anstey NM, Buffet PA, Poespoprodjo JR, Yeo TW, White NJ, et al. The anaemia of Plasmodium vivax malaria. Malar J 2012;11:135. https://doi.org/10.1186/1475-2875-11-135.
Ellis JM, Stubbs TH, Young MD. Some Characteristics of Foreign Vivax Malaria Induced in Neurosyphilitic Patients 1,2. Am J Trop Med Hyg 1947;s1-27:585–96. https://doi.org/10.4269/ajtmh.1947.s1-27.585.
Galinski MR, Medina CC, Ingravallo P, Barnwell JW. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell 1992;69:1213–26. https://doi.org/10.1016/0092-8674(92)90642-P.
Golassa L, Amenga-Etego L, Lo E, Amambua-Ngwa A. The biology of unconventional invasion of Duffy-negative reticulocytes by Plasmodium vivax and its implication in malaria epidemiology and public health. Malar J 2020;19. https://doi.org/10.1186/s12936-020-03372-9.
Gonzalez L, Vega J, Ramirez JL, Bedoya G, Carmona-Fonseca J, Maestre A. Relationship between genotypes of the Duffy blood groups and malarial infection in different ethnic groups of Choco, Colombia. Colomb Med (Cali) 2012;43:189–95.
Gunalan K, Lo E, Hostetler JB, Yewhalaw D, Mu J, Neafsey DE, et al. Role of Plasmodium vivax Duffy-binding protein 1 in invasion of Duffy-null Africans. Proceedings of the National Academy of Sciences 2016;113:6271–6. https://doi.org/10.1073/pnas.1606113113.
Gunalan K, Niangaly A, Thera MA, Doumbo OK, Miller LH. Plasmodium vivax Infections of Duffy-Negative Erythrocytes: Historically Undetected or a Recent Adaptation? Trends Parasitol 2018;34. https://doi.org/10.1016/j.pt.2018.02.006.
Gupta S, Singh S, Popovici J, Roesch C, Shakri AR, Guillotte-Blisnick M, et al. Targeting a Reticulocyte Binding Protein and Duffy Binding Protein to Inhibit Reticulocyte Invasion by Plasmodium vivax. Sci Rep 2018;8:10511. https://doi.org/10.1038/s41598-018-28757-4.
Han J-H, Cheng Y, Muh F, Ahmed MA, Cho J-S, Nyunt MH, et al. Inhibition of parasite invasion by monoclonal antibody against epidermal growth factor-like domain of Plasmodium vivax merozoite surface protein 1 paralog. Sci Rep 2019;9:3906. https://doi.org/10.1038/s41598-019-40321-2.
Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, et al. The global distribution of the Duffy blood group. Nat Commun 2011;2:266. https://doi.org/10.1038/ncomms1265.
INS-Boletin epidemiologico semanal. Semana epidemiológica 52; 24 al 30 de diciembre de 2023. 2023.
Kadekoppala M, Holder AA. Merozoite surface proteins of the malaria parasite: The MSP1 complex and the MSP7 family. Int J Parasitol 2010;40:1155–61. https://doi.org/10.1016/j.ijpara.2010.04.008.
Kanjee U, Rangel GW, Clark MA, Duraisingh MT. Molecular and cellular interactions defining the tropism of Plasmodium vivax for reticulocytes. Curr Opin Microbiol 2018;46. https://doi.org/10.1016/j.mib.2018.10.002.
Kar S, Sinha A. Plasmodium vivax Duffy Binding Protein-Based Vaccine: a Distant Dream. Front Cell Infect Microbiol 2022;12:916702. https://doi.org/10.3389/fcimb.2022.916702.
King CL, Adams JH, Xianli J, Grimberg BT, McHenry AM, Greenberg LJ, et al. Fy a /Fy b antigen polymorphism in human erythrocyte Duffy antigen affects susceptibility to Plasmodium vivax malaria. Proceedings of the National Academy of Sciences 2011;108:20113–8. https://doi.org/10.1073/pnas.1109621108.
Kwiatkowski , Dominic P. How Malaria Has Affected the Human Genome and What Human Genetics Can Teach Us about Malaria . The American Journal of Human Genetics n.d.;77.
Lo E, Hostetler JB, Yewhalaw D, Pearson RD, Hamid MMA, Gunalan K, et al. Frequent expansion of Plasmodium vivax Duffy Binding Protein in Ethiopia and its epidemiological significance. PLoS Negl Trop Dis 2019;13:e0007222. https://doi.org/10.1371/journal.pntd.0007222.
Loy DE, Liu W, Li Y, Learn GH, Plenderleith LJ, Sundararaman SA, et al. Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. Int J Parasitol 2017;47. https://doi.org/10.1016/j.ijpara.2016.05.008.
Malleret B, Li A, Zhang R, Tan KSW, Suwanarusk R, Claser C, et al. Plasmodium vivax: restricted tropism and rapid remodeling of CD71-positive reticulocytes. Blood 2015;125:1314–24. https://doi.org/10.1182/blood-2014-08-596015.
Martínez Santander C ACCCSLCVHRAK. Malaria: Un problema de alta prevalencia en habitantes de regiones tropicales de Latinoamérica. Ateneo 2023;25(2):110–28.
Ménard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci U S A 2010;107:5967–71. https://doi.org/10.1073/pnas.0912496107.
Menard D, Chan ER, Benedet C, Ratsimbasoa A, Kim S, Chim P, et al. Whole Genome Sequencing of Field Isolates Reveals a Common Duplication of the Duffy Binding Protein Gene in Malagasy Plasmodium vivax Strains. PLoS Negl Trop Dis 2013;7:e2489. https://doi.org/10.1371/journal.pntd.0002489.
Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de Sousa B, et al. Duffy Negative Antigen Is No Longer a Barrier to Plasmodium vivax – Molecular Evidences from the African West Coast (Angola and Equatorial Guinea). PLoS Negl Trop Dis 2011;5:e1192. https://doi.org/10.1371/journal.pntd.0001192.
Mercereau-Puijalon O, Ménard D. Plasmodium vivax and the Duffy antigen: A paradigm revisited. Transfusion Clinique et Biologique 2010;17:176–83. https://doi.org/10.1016/j.tracli.2010.06.005.
Miller LH, Mason SJ, Clyde DF, McGinniss MH. The Resistance Factor to Plasmodium vivax in Blacks. The Duffy Blood Group Genotype, FyFy. N Engl J Med 1976;295.
Molina-Franky J, Reyes C, Picón Jaimes YA, Kalkum M, Patarroyo MA. The Black Box of Cellular and Molecular Events of Plasmodium vivax Merozoite Invasion into Reticulocytes. Int J Mol Sci 2022;23. https://doi.org/10.3390/ijms232314528.
Molina-Franky J, Reyes C, Picón Jaimes YA, Kalkum M, Patarroyo MA. The Black Box of Cellular and Molecular Events of Plasmodium vivax Merozoite Invasion into Reticulocytes. Int J Mol Sci 2022;23. https://doi.org/10.3390/ijms232314528.
Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, et al. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis 2009;9. https://doi.org/10.1016/S1473-3099(09)70177-X.
Niangaly A, Gunalan K, Ouattara A, Coulibaly D, Sá JM, Adams M, et al. Plasmodium vivax Infections over 3 Years in Duffy Blood Group Negative Malians in Bandiagara, Mali. American Journal of Tropical Medicine and Hygiene 2017;97. https://doi.org/10.4269/ajtmh.17-0254.
Organización Mundial de la Salud/ Organización Panamericana de la Salud. Malaria. Https://WwwWhoInt/Es/News-Room/Fact-Sheets/Detail/Malaria 2023.
Pachebat JA, Kadekoppala M, Grainger M, Dluzewski AR, Gunaratne RS, Scott-Finnigan TJ, et al. Extensive proteolytic processing of the malaria parasite merozoite surface protein 7 during biosynthesis and parasite release from erythrocytes. Mol Biochem Parasitol 2007;151:59–69. https://doi.org/10.1016/j.molbiopara.2006.10.006.
Pearson RD, Amato R, Auburn S, Miotto O, Almagro-Garcia J, Amaratunga C, et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet 2016;48:959–64. https://doi.org/10.1038/ng.3599.
Picón-Jaimes YA, Lozada-Martinez ID, Buelvas MCF, Sarmiento AFA, Baez GAS, Erazo DYN, et al. Evolution of Plasmodium vivax and resistance patterns for infection based on Duffy genotype and phenotype. Infezioni in Medicina 2023;31:350–8. https://doi.org/10.53854/liim-3103-8.
Poirier P, Doderer-Lang C, Atchade PS, Lemoine J-P, de l’Isle M-LC, Abou-bacar A, et al. The hide and seek of Plasmodium vivax in West Africa: report from a large-scale study in Beninese asymptomatic subjects. Malar J 2016;15:570. https://doi.org/10.1186/s12936-016-1620-z.
Popovici J, Roesch C, Rougeron V. The enigmatic mechanisms by which Plasmodium vivax infects Duffy-negative individuals. PLoS Pathog 2020;16. https://doi.org/10.1371/journal.ppat.1008258.
Roesch C, Popovici J, Bin S, Run V, Kim S, Ramboarina S, et al. Genetic diversity in two Plasmodium vivax protein ligands for reticulocyte invasion. PLoS Negl Trop Dis 2018;12:e0006555. https://doi.org/10.1371/journal.pntd.0006555.
Roobsoong W, Tharinjaroen CS, Rachaphaew N, Chobson P, Schofield L, Cui L, et al. Improvement of culture conditions for long-term in vitro culture of Plasmodium vivax. Malar J 2015;14:297. https://doi.org/10.1186/s12936-015-0815-z.
Russell B, Suwanarusk R, Borlon C, Costa FTM, Chu CS, Rijken MJ, et al. A reliable ex vivo invasion assay of human reticulocytes by Plasmodium vivax. Blood 2011;118:e74–81. https://doi.org/10.1182/blood-2011-04-348748.
Russo G, Faggioni G, Paganotti GM, Djeunang Dongho GB, Pomponi A, De Santis R, et al. Molecular evidence of Plasmodium vivax infection in Duffy negative symptomatic individuals from Dschang, West Cameroon. Malar J 2017;16:74. https://doi.org/10.1186/s12936-017-1722-2.
Tyagi K, Hossain ME, Thakur V, Aggarwal P, Malhotra P, Mohmmed A, et al. Plasmodium vivax Tryptophan Rich Antigen PvTRAg36.6 Interacts with PvETRAMP and PvTRAg56.6 Interacts with PvMSP7 during Erythrocytic Stages of the Parasite. PLoS One 2016;11:e0151065. https://doi.org/10.1371/journal.pone.0151065.
Young MD, Eyles DE, Burgess RW, Jeffery GM. Experimental Testing of the Immunity of Negroes to Plasmodium vivax. J Parasitol 1955;41:315. https://doi.org/10.2307/3274214.
Descargas
Publicado
Cómo citar
Número
Sección
Licencia
Derechos de autor 2025 Hechos Microbiológicos

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-CompartirIgual 4.0.