Gliptins vs. Milk-derived Dipeptidyl-Peptidase IV Inhibiting Biopeptides: Physicochemical Characterization and Pharmacokinetic Profiling
DOI:
https://doi.org/10.17533/udea.vitae.v28n3a346531Keywords:
Bioactive peptides, Dipeptidyl-Peptidase IV inhibitors, Type 2 Diabetes Mellitus, PharmacokineticsAbstract
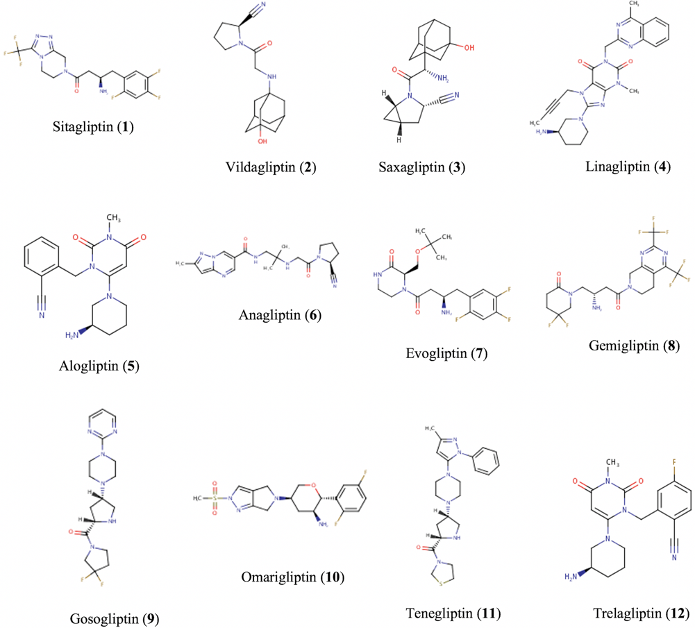
Background: Milk-derived biopeptides have reported in vitro dipeptidyl-peptidase IV (DPP-IV) inhibition, suggesting a glycemic-regulatory effect in Type 2 Diabetes Mellitus (T2DM). Nonetheless, the therapeutic application of these nutraceuticals is limited by the scarcity of knowledge regarding their pharmacokinetic profile. Objective: This study aimed to characterize and assess the pharmacokinetics of milk-derived biopeptides. Through an in silico comparative analysis with gliptins, we expected to identify enhanced properties in food-hydrolysates and suitable DPP-IV inhibiting peptides as candidates for T2DM therapy. Methods: A comparison between gliptins and biopeptides was conducted based on in silico evaluation of drug-likeness, physicochemical properties, pharmacokinetics, and synthetic accessibility. Suitable target proteins for gastrointestinal-absorbable biopeptides were determined as well. Data collection was performed on SwissADME, ADMETlab, DrugBank, SwissTargetPrediction, ChemDes, and BIOPEP-UWM platforms. Statistical analysis was carried out using a one-way ANOVA test. Results: Drug-likeness compliance showed no significant difference between gliptins and biopeptides (p>0.05) in three out of nine assessed rules, though gastrointestinal-absorbable biopeptides exhibited no significant difference with gliptins in five drug-likeness guidelines. The physicochemical evaluation revealed a significant difference (p<0.05) between both groups, with peptides exhibiting enhanced solubility, flexibility, and polarity. Nine out of thirty-six assessed biopeptides reported being likely gastrointestinal-absorbable molecules, from which six displayed ≥30% predicted bioavailability, two reported CYP450 interactions, and all were determined to be blood confined. Biopeptides showed a slightly lower clearance than gliptins yet counteracted by a significantly lower half-life. Moreover, synthetic accessibility scores indicated higher synthetic ease for biopeptides. In addition, absorbable bioactive peptides reported a considerable binding affinity to DPP-IV and Calpain-I. Conclusions: Compared to gliptins, gastrointestinal-absorbable biopeptides exhibit superior physicochemical properties (higher solubility, flexibility, and polarity), lesser CYP450 interactions, higher synthetic ease, and some reported an important affinity for DPP-IV and Calpain-I. Only a small fraction of milk-derived biopeptides are suitable drug-like compounds and feasible candidates for T2DM therapy; yet, testing their therapeutic potency remains subject to further studies.
Downloads
References
Cole JB, Florez JC. Genetics of diabetes mellitus and diabetes complications. Nat Rev Nephrol. 2020;16(7):377–90. DOI: https://doi.org/10.1038/s41581-020-0278-5
Rieddle MC, Ahmann AJ. Therapeutics of Type 2 Diabetes Mellitus. In: Williams Textbook of Endocrinology. 14th ed. Elsevier; 2020. p. 1386.
Doumas M, Imprialos K, Stavropoulos K, Athyros VG. Pharmacological Management of Type 2 Diabetes Complications. Curr Vasc Pharmacol. 2020;18(2):101–3. DOI: https://doi.org/10.2174/157016111802200101155519
Mockus I, Trujillo M. Obesidad y Enfermedades Asociadas, 1st ed., Ismena M, Martha T, editors. Universidad Nacional de Colombia; 2013. 230 p.
Paschou SA, Siasos G, Bletsa E, Stampouloglou PK, Oikonomou E, Antonopoulos AS, et al. The Effect of DPP-4i on Endothelial Function and Arterial Stiffness in Patients with Type 2 Diabetes: A Systematic Review of Randomized Placebo-controlled Trials. Curr Pharm Des. 2020;26(46):5980–7. DOI: https://doi.org/10.2174/1381612826666200417153241
Sachdeva V, Roy A, Bharadvaja N. Current Prospects of Nutraceuticals: A Review. Curr Pharm Biotechnol. 2020;21(10):884–96. DOI: https://doi.org/10.2174/1389201021666200130113441
Premi M, Bansal V. Nutraceuticals for Management of Metabolic Disorders. In: Treating endocrine and metabolic disorders with herbal medicines. 2021. p. 298–320.
Jao C-L, Hung C-C, Tung Y-S, Lin P-Y, Chen M-C, Hsu K-C. The development of bioactive peptides from dietary proteins as a dipeptidyl peptidase IV inhibitor for the management of type 2 diabetes. BioMedicine. 2015;5(3):14. DOI: https://doi.org/10.7603/s40681-015-0014-9
Patil P, Mandal S, Tomar SK, Anand S. Food protein-derived bioactive peptides in management of type 2 diabetes. Eur J Nutr. 2015;54(6):863–80. DOI: https://doi.org/10.1007/s00394-015-0974-2
Acquah C, Dzuvor CK, Tosh S, Agyei D. Anti-diabetic effects of bioactive peptides: recent advances and clinical implications. Crit Rev Food Sci Nutr. 2020;1–14. DOI: https://doi.org/10.1080/10408398.2020.1851168
Iwaniak A, Minkiewicz P, Darewicz M, Hrynkiewicz M. Food protein-originating peptides as tastants - Physiological, technological, sensory, and bioinformatic approaches. Food Res Int. 2016;89:27–38. DOI: https://doi.org/10.1016/j.foodres.2016.08.010
Dong J, Wang N-N, Yao Z-J, Zhang L, Cheng Y, Ouyang D, et al. ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J Cheminformatics. 2018;10(1):29. DOI: https://doi.org/10.1186/s13321-018-0283-x
Barrero JA, Cruz CM, Casallas J, Vásquez JS. Evaluación in silico de péptidos bioactivos derivados de la digestión de proteínas presentes en la leche de bovino (B.taurus), oveja (O.aries), cabra (C.hircus) y búfalo (B.bubalis). TecnoLógicas. 2020;50(24). DOI: https://doi.org/10.22430/22565337.1731
Nongonierma AB, Mooney C, Shields DC, FitzGerald RJ. Inhibition of dipeptidyl peptidase IV and xanthine oxidase by amino acids and dipeptides. Food Chem. 2013;141(1):644–53. DOI: https://doi.org/10.1016/j.foodchem.2013.02.115
Nongonierma AB, FitzGerald RJ. An in silico model to predict the potential of dietary proteins as sources of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides. Food Chem. 2014;165:489–98. DOI: https://doi.org/10.1016/j.foodchem.2014.05.090
Cheung HS, Wang FL, Ondetti MA, Sabo EF, Cushman DW. Binding of peptide substrates and inhibitors of angiotensin-converting enzyme. Importance of the COOH-terminal dipeptide sequence. J Biol Chem. 1980;255(2):401–7. https://pubmed.ncbi.nlm.nih.gov/6243277/
Lan VTT, Ito K, Ohno M, Motoyama T, Ito S, Kawarasaki Y. Analyzing a dipeptide library to identify human dipeptidyl peptidase IV inhibitor. Food Chem. 2015;175:66–73. DOI: https://doi.org/10.1016/j.foodchem.2014.11.131
Nongonierma AB, FitzGerald RJ. Susceptibility of milk protein-derived peptides to dipeptidyl peptidase IV (DPP-IV) hydrolysis. Food Chem. 2014;145:845–52. DOI: https://doi.org/10.1016/j.foodchem.2013.08.097
Nongonierma AB, FitzGerald RJ. Inhibition of dipeptidyl peptidase IV (DPP-IV) by proline containing casein-derived peptides. J Funct Foods. 2013;5(4):1909–17. DOI: https://doi.org/10.1016/j.jff.2013.09.012
Silveira ST, Martínez D, Recio I, Hernández B. Dipeptidyl peptidase-IV inhibitory peptides generated by tryptic hydrolysis of a whey protein concentrate rich in β-lactoglobulin. Food Chem. 2013;141(2):1072–7. DOI: https://doi.org/10.1016/j.foodchem.2013.03.056
Gallego M, Aristoy M-C, Toldrá F. Dipeptidyl peptidase IV inhibitory peptides generated in Spanish dry-cured ham. Meat Sci. 2014;96(2):757–61. DOI: https://doi.org/10.1016/j.meatsci.2013.09.014
Dong J, Cao D-S, Miao H-Y, Liu S, Deng B-C, Yun Y-H, et al. ChemDes: an integrated web-based platform for molecular descriptor and fingerprint computation. J Cheminformatics. 2015;7(1):60. DOI: https://doi.org/10.1186/s13321-015-0109-z
Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(1):42717. DOI: https://doi.org/10.1038/srep42717
Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–64. DOI: https://doi.org/10.1093/nar/gkz382
Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–82. DOI: https://doi.org/10.1093/nar/gkx1037
Minkiewicz, Iwaniak, Darewicz. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int J Mol Sci. 2019;20(23):5978. DOI: https://doi.org/10.3390/ijms20235978
Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods. 2000;44(1):235–49. DOI: https://doi.org/10.1016/s1056-8719(00)00107-6
Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J Med Chem. 2002;45(12):2615–23. DOI: https://doi.org/10.1021/jm020017n
Ghose AK, Viswanadhan VN, Wendoloski JJ. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J Comb Chem. 1999;1(1):55–68. DOI: https://doi.org/10.1021/cc9800071
Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–41. DOI: https://doi.org/10.1016/j.ddtec.2004.11.007
Oprea TI. Property distribution of drug-related chemical databases. J Comput Aided Mol Des. 2000;14(3):251–64. DOI: https://doi.org/10.1023/A:1008130001697
Gupta M, Lee HJ, Barden CJ, Weaver DF. The Blood–Brain Barrier (BBB) Score. J Med Chem. 2019;62(21):9824–36. DOI: https://doi.org/10.1021/acs.jmedchem.9b01220
Egan WJ, Merz, KM, Baldwin JJ. Prediction of Drug Absorption Using Multivariate Statistics. J Med Chem. 2000;43(21):3867–77. DOI: https://doi.org/10.1021/jm000292e
Muegge I, Heald SL, Brittelli D. Simple Selection Criteria for Drug-like Chemical Matter. J Med Chem. 2001;44(12):1841–6. DOI: https://doi.org/10.1021/jm015507e
Varma MV, Obach RS, Rotter C, Miller HR, Chang G, Steyn SJ, et al. Physicochemical Space for Optimum Oral Bioavailability: Contribution of Human Intestinal Absorption and First-Pass Elimination. J Med Chem. 2010;53(3):1098–108. DOI: https://doi.org/10.1021/jm901371v
Daina A, Zoete V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem. 2016;11(11):1117-21. DOI: https://doi.org/10.1002/cmdc.201600182
Tian S, Li Y, Wang J, Zhang J, Hou T. ADME Evaluation in Drug Discovery. 9. Prediction of Oral Bioavailability in Humans Based on Molecular Properties and Structural Fingerprints. Mol Pharm. 2011;8(3):841–51. DOI: https://doi.org/10.1021/mp100444g
Tian S, Wang J, Li Y, Li D, Xu L, Hou T. The application of in silico drug-likeness predictions in pharmaceutical research. Adv Drug Deliv Rev. 2015;86:2–10. DOI: https://doi.org/ 10.1016/j.addr.2015.01.009
Ou-Yang S, Lu J, Kong X, Liang Z, Luo C, Jiang H. Computational drug discovery. Acta Pharmacol Sin. 2012;33(9):1131–40. DOI: https://doi.org/10.1038/aps.2012.109
Loureiro DRP, Soares JX, Costa JC, Magalhães ÁF, Azevedo CMG, Pinto MMM, et al. Structures, Activities and Drug-Likeness of Anti-Infective Xanthone Derivatives Isolated from the Marine Environment: A Review. Molecules. 2019;24(2):243. DOI: https://doi.org/10.3390/molecules24020243
Hitchcock SA. Blood–brain barrier permeability considerations for CNS-targeted compound library design. Curr Opin Chem Biol. 2008;12(3):318–23. DOI: https://doi.org/10.1016/j.cbpa.2008.03.019
Jia C-Y, Li J-Y, Hao G-F, Yang G-F. A drug-likeness toolbox facilitates ADMET study in drug discovery. Drug Discov Today. 2020;25(1):248–58. DOI: https://doi.org/10.1016/j.drudis.2019.10.014
J Ji D, Xu M, Udenigwe CC, Agyei D. Physicochemical characterisation, molecular docking, and drug-likeness evaluation of hypotensive peptides encrypted in flaxseed proteome. Curr Res Food Sci. 2020;3:41–50. DOI: https://doi.org/10.1016/j.crfs.2020.03.001
Meanwell NA. Improving Drug Candidates by Design: A Focus on Physicochemical Properties As a Means of Improving Compound Disposition and Safety. Chem Res Toxicol. 2011 Sep 19;24(9):1420–56. DOI: https://doi.org/10.1021/tx200211v
Das T, Mehta CH, Nayak UY. Multiple approaches for achieving drug solubility: an in silico perspective. Drug Discov Today. 2020;25(7):1206–12. DOI: https://doi.org/10.1016/j.drudis.2020.04.016
Caron G, Digiesi V, Solaro S, Ermondi G. Flexibility in early drug discovery: focus on the beyond-Rule-of-5 chemical space. Drug Discov Today. 2020;25(4):621–7. DOI: https://doi.org/10.1016/j.drudis.2020.01.012
Testa B, van de Waterbeemd H, Folkers G, Guy R. Pharmacokinetic Optimization in Drug Research: Biological, Physicochemical, and Computational Strategies [Internet]. 1st ed. Wiley; 2001 [cited 2021 May 22]. Available from: https://onlinelibrary.wiley.com/doi/book/10.1002/9783906390437
Acquah C, Stefano ED, Udenigwe CC. Role of hydrophobicity in food peptide functionality and bioactivity. J Food Bioact. 2018;4(1):88–98. DOI: https://doi.org/10.31665/JFB.2018.4164
Shultz MD. Two Decades under the Influence of the Rule of Five and the Changing Properties of Approved Oral Drugs. J Med Chem. 2019;62(4):1701–14. DOI: https://doi.org/10.1021/acs.jmedchem.8b00686
Alam A, Kowal J, Broude E, Roninson I, Locher KP. Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science. 2019;363(6428):753–6. DOI: https://doi.org/10.1126/science.aav7102
Di L, Kerns EH. Drug-Like Properties - Concepts, Structure Design and Methods from ADME to Toxicity Optimization [Internet]. 2nd ed. Elsevier; 2016 [cited 2021 May 2]. 580 p. Available from: https://linkinghub.elsevier.com/retrieve/pii/C2013018378X
Mulvihill EE, Varin EM, Gladanac B, Campbell JE, Ussher JR, Baggio LL, et al. Cellular Sites and Mechanisms Linking Reduction of Dipeptidyl Peptidase-4 Activity to Control of Incretin Hormone Action and Glucose Homeostasis. Cell Metab. 2017;25(1):152–65. DOI: https://doi.org/10.1016/j.cmet.2016.10.007
Kim MT, Sedykh A, Chakravarti SK, Saiakhov RD, Zhu H. Critical Evaluation of Human Oral Bioavailability for Pharmaceutical Drugs by Using Various Cheminformatics Approaches. Pharm Res. 2014;31(4):1002–14. DOI: https://doi.org/10.1007/s11095-013-1222-1
Isvoran A, Louet M, Vladoiu DL, Craciun D, Loriot M-A, Villoutreix BO, et al. Pharmacogenomics of the cytochrome P450 2C family: impacts of amino acid variations on drug metabolism. Drug Discov Today. 2017;22(2):366–76. DOI: https://doi.org/10.1016/j.drudis.2016.09.015
Deodhar M, Rihani SBA, Darakjian L, Turgeon J, Michaud V. Assessing the Mechanism of Fluoxetine-Mediated CYP2D6 Inhibition. Pharmaceutics. 2021;13(2):148. DOI: https://doi.org/10.3390/pharmaceutics13020148
Smith DA, Beaumont K, Maurer TS, Di L. Relevance of Half-Life in Drug Design. J Med Chem. 2018;61(10):4273–82. DOI: https://doi.org/10.1021/acs.jmedchem.7b00969
Mignani S, Rodrigues J, Tomas H, Jalal R, Singh PP, Majoral J-P, et al. Present drug-likeness filters in medicinal chemistry during the hit and lead optimization process: how far can they be simplified? Drug Discov Today. 2018;23(3):605–15. DOI: https://doi.org/10.1016/j.drudis.2018.01.010
Teague SJ, Davis AM, Leeson PD, Oprea T. The Design of Leadlike Combinatorial Libraries. Angew Chem Int Ed. 1999;38(24):3743–8. DOI: https://doi.org/10.1002/(SICI)1521-3773(19991216)38:24<3743::AID-ANIE3743>3.0.CO;2-U
Ertl P, Schuffenhauer A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J Cheminformatics. 2009;1(1):8. DOI: https://doi.org/10.1186/1758-2946-1-8
Azad I, Nasibullah M, Khan T, Hassan F, Akhter Y. Exploring the novel heterocyclic derivatives as lead molecules for design and development of potent anticancer agents. J Mol Graph Model. 2018;81(1):211–28. DOI: https://doi.org/10.1016/j.jmgm.2018.02.013
Iwaniak A, Minkiewicz P, Pliszka M, Mogut D, Darewicz M. Characteristics of Biopeptides Released In Silico from Collagens Using Quantitative Parameters. Foods. 2020;9(7):965. DOI: https://doi.org/10.3390/foods9070965
Kyselova A, Elgheznawy A, Wittig I, Heidler J, Mann AW, Ruf W, et al. Platelet-derived calpain cleaves the endothelial protease-activated receptor 1 to induce vascular inflammation in diabetes. Basic Res Cardiol. 2020;115(6):75. DOI: https://doi.org/10.1007/s00395-020-00833-9
Dókus LE, Yousef M, Bánóczi Z. Modulators of calpain activity: inhibitors and activators as potential drugs. Expert Opin Drug Discov. 2020;15(4):471–86. DOI: https://doi.org/10.1080/17460441.2020.1722638
Donkor IO. An update on the therapeutic potential of calpain inhibitors: a patent review. Expert Opin Ther Pat. 2020;30(9):659–75. DOI: https://doi.org/10.1080/13543776.2020.1797678
Andrade EL, Bento AF, Cavalli J, Oliveira SK, Freitas CS, Marcon R, et al. Non-clinical studies required for new drug development - Part I: early in silico and in vitro studies, new target discovery and validation, proof of principles and robustness of animal studies. Braz J Med Biol Res. 2016;49(11):e5644. DOI: https://doi.org/10.1590/1414-431X20165644
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2021 Jorge Andrés Barrero, Fabio Cabrera, Claudia Marcela Cruz

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Copyright Notice and Open Access Statement
The Journal Vitae works under the Open Access license, and the published manuscripts remain available for the public, both on the Journal's website and in databases, under the Creative Commons license, "Noncommercial Attribution" and "Share alike" systems, adopted in Colombia. Hence, when the authors agree to publish in the Journal Vitae, they will not have the right to economic retributions on publications and reproductions through different diffusion media. The documents are freely available to the internet public, permitting users to read, download, copy, distribute, print, search, or link to the full texts and pass them as data to software. The only constraint on reproduction and distribution, should be to give authors control over the integrity of their work and the right to be appropriately acknowledged and cited.
Authors declare that:
-
They are the intellectual property owners and are responsible for all the information stated in the article.
-
This manuscript has not been submitted or published in other printed or digital media. They accept the responsibility for the judgments, opinions, and points of view expressed in the published article and, therefore, they exonerate Universidad de Antioquia and Journal Vitae from any process.
-
They exempt Universidad de Antioquia and Journal Vitae from settling conflicts or disputes related to the authorship of the referred article.
-
They accept the revision of the original manuscript by suitable personnel, and they bind themselves to perform the corrections appointed or suggested by the assessors.
-
Therefore, they know the editorial process and will not bind the Editorial Board of the Journal to assume any obligations regarding the volume and issue in which the article is published.
-
They transfer the rights of publication, reprinting, and distribution of the article from the moment of its approval, in print and digital format, without the right to economic rewards, and under the licensing conditions considered relevant by Journal Vitae.
-
They fully authorize Universidad de Antioquia and Journal Vitae to submit the published material to the diverse databases and indexing systems where the Journal can be found to comply with the requirements of the regulatory authorities to maintain the national classification of journals.
-
They will assume the article publication costs established for the current issue, and they will make the payment as soon as they are informed about the volume and the issue in which the final version of the article is published.
-
After the article is published, you can share digital or printed copies in a noncommercial manner. You will be able to use the paper in your institution or company for educational or research purposes, including the use in course programs.
Conflict of interest: Authors are responsible for recognizing and disclosing any financial or other benefits that could be perceived to bias their work, acknowledging all financial support and any personal connections with potential sponsors. Examples of such conflicts include receiving research funds or honoraria, serving on advisory boards, stock ownership, or employment and consulting arrangements. Authors without such connections should clearly state that they have no financial support or personal relationships that could be perceived to bias their work. All conflicts of interest should be disclosed on the author's identification page of the manuscript.









